Before we knew it, we’ve reached the last series of our data tutorials. Each chapter is independent. If you want to learn the previous tutorials, you can click on the links below.
10 Efficient Data Cleaning Methods Every Python Crawler Should Know(4) Bloom Filter
10 Efficient Data Cleaning Methods Every Python Crawler Should Know(3)
10 Efficient Data Cleaning Methods Every Python Crawler Should Know(2)
10 Efficient Data Cleaning Methods Every Python Crawler Should Know(1)
In the field of data processing, pandas is undoubtedly one of the most powerful data processing libraries in the Python ecosystem. Mastering it will give you an edge, and you might even be able to ditch Excel.
9. pandas data processing
pandas installation:
pip install pandasFor the data process, we’ll refer to the read_html method in the first tutorial:10 Efficient Data Cleaning Methods Every Python Crawler Should Know(1)
import pandas as pd
table_MN = pd.read_html('https://en.wikipedia.org/wiki/Minnesota')Multiple table data are read, and the type is a list. Check how many tables there are.
print(f'Total tables: {len(table_MN)}')Total tables: 38If you input the title of a table, you can also directly obtain the corresponding table data.
table_MN = pd.read_html('https://en.wikipedia.org/wiki/Minnesota', match='Election results from statewide races')
len(table_MN)df = table_MN[0]
df.head()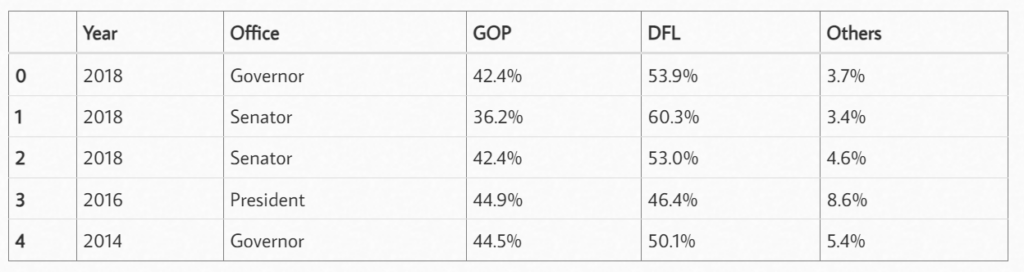
Check the data type of each column of data:
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 24 entries, 0 to 23
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Year 24 non-null int64
1 Office 24 non-null object
2 GOP 24 non-null object
3 DFL 24 non-null object
4 Others 24 non-null object
dtypes: int64(1), object(4)
memory usage: 1.1+ KBIf we want to conduct any analysis, we need to convert the “GOP”, “DFL”, and “Others” columns into numerical data.
If we use the following statement to convert “GOP” to numbers:
df['GOP'].astype('float')Then an error will occur:
ValueError: could not convert string to float: '42.4%'Since the “%” symbol cannot be converted to a number, we need to remove it before conversion.
df['GOP'].replace({'%':''}, regex=True).astype('float')In this way, we get the correct data.
0 42.4
1 36.2
2 42.4
3 44.9
<...>
21 63.3
22 49.1
23 31.9
Name: GOP, dtype: float64If you want to convert all the other columns in the table to numbers:
df = df.replace({'%': ''}, regex=True)
df[['GOP', 'DFL', 'Others']] = df[['GOP', 'DFL', 'Others']].apply(pd.to_numeric)
df.info()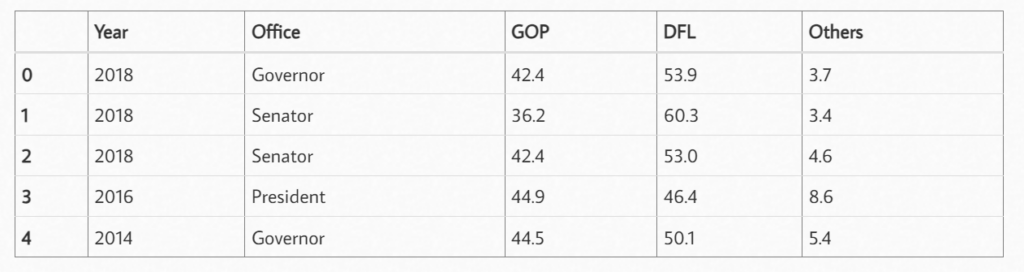
In this way, all the “%” symbols in the table are removed.
Handling Missing Values
Missing values may affect the analysis results. Pandas provides methods such as dropna() and fillna() to handle missing data.
- Detecting Missing Values
isnull()is a function in Pandas used to detect missing values. It returns a boolean DataFrame, whereTrueindicates that the position is a missing value (NaN), andFalseindicates that the position has valid data.
import pandas as pd
df = pd.DataFrame({
'A': [1, 2, None, 4],
'B': [None, 2, 3, 4],
'C': [1, None, None, 4]
})
# Detect missing values and display them cell by cell. True/False
print(df.isnull())The output result:
A B C
0 False True False
1 False False True
2 True False True
3 False False FalseIn the above output:
- The first row indicates that the first value in column
Ais valid (False), while the first value in columnBis missing (True). - The second row indicates that the second value in column
Cis missing (True).
isnull().sum() Usage:
isnull().sum() is used to count the number of missing values in each column. isnull() returns a boolean DataFrame, and sum() can sum up the True (missing values) by column, thus obtaining the number of missing values in each column.
Count the number of missing values in each column.
print(df.isnull().sum())The output result:
A 1
B 1
C 2
dtype: int64- Column
Ahas 1 missing value. - Column
Bhas 1 missing value. - Column
Chas 2 missing values.
Deleting Missing Values: dropna()
# delete NaN row
df_cleaned = df.dropna()
# delete NaN column
df_cleaned_cols = df.dropna(axis=1) Handling Duplicate Values
The data may contain duplicate rows. Use duplicated() and drop_duplicates() to handle them.
**duplicated() method
duplicated() is used to detect duplicate rows in a DataFrame. It returns a boolean Series, where each element indicates whether the corresponding row is a duplicate row (the first occurrence of a row will be marked as False, and subsequent duplicate rows will be marked as True).
import pandas as pd
df = pd.DataFrame({
'A': [1, 2, 2, 4],
'B': [5, 6, 6, 8]
})
# detect duplicated row
print(df.duplicated())Output:
0 False
1 False
2 True
3 False
dtype: bool- Row 0 is
Falsebecause it is the first occurrence of[1, 5]. - Row 1 is
Falsebecause it is the first occurrence of[2, 6]. - Row 2 is
Truebecause it is a duplicate of[2, 6], which is a duplicate of row 1. - Row 3 is
Falsebecause it is the first occurrence of[4, 8].
drop_duplicates() Method
drop_duplicates() is used to delete duplicate rows and only keep unique rows. It returns a new DataFrame with duplicate rows removed.
# delete duplicated row
df_no_duplicates = df.drop_duplicates()
print(df_no_duplicates)Output:
A B
0 1 5
1 2 6
3 4 8pd.to_datetime()
It is used to convert data such as strings, integers, and timestamps into Pandas’ datetime type, which facilitates the processing and analysis of time series.
str.strip()
It is used to remove leading and trailing whitespace characters (including spaces, tabs, newlines, etc.) from a string.
str.lower()
It is used to convert a string to lowercase letters.
str.replace()
It is used to replace certain characters or substrings in a string.
import pandas as pd
df = pd.DataFrame({
'A': ['1', '2', '3', '4'],
'B': ['2021-01-01', '2021-02-01', '2021-03-01', '2021-04-01'],
'C': [' hello ', ' world ', ' old_value ', ' new_value ']
})
# data type convertion
df['A'] = df['A'].astype(float) # covnert to float
df['B'] = pd.to_datetime(df['B'], errors='coerce') # convert to datetime
# string type convertion
df['C'] = df['C'].str.strip() # remove space
df['C'] = df['C'].str.lower() # lower case
df['C'] = df['C'].replace({'old_value': 'new_value'}) # replace value
print(df)Output result:
A B C
0 1.0 2021-01-01 hello
1 2.0 2021-02-01 world
2 3.0 2021-03-01 new_value
3 4.0 2021-04-01 new_valueString Regular Expressions
String processing is a very common scenario. Sometimes, you need to perform regular expression operations on an entire column based on a certain field, and DataFrame also supports this.
Using regular expressions to extract information
df['phone_area'] = df['phone'].str.extract(r'\((\d{3})\)')Vectorized string concatenation
df['name'] = df['first_name'].str.cat(df['last_name'], sep=' ')Fuzzy matching
Use fuzzywuzzy for fuzzy matching.
from fuzzywuzzy import fuzz
df['similarity'] = df.apply(lambda x: fuzz.ratio(x['name1'], x['name2']), axis=1)(If you get a prompt saying that fuzzywuzzy cannot be found, you can install it using pip: pip install fuzzywuzzy)
High-performance Data Merging
For example, if you have two tables, one for Company A and the other for Company B, you need to merge the two tables into one.
Or if one table contains users’ basic information and the other contains sales lists, you often need to output the two tables as one large table to calculate the gender of each order purchaser.
There are several commonly used methods for data merging.
Fast Merging Based on Index
df1.join(df2, how='left')- Example code
import pandas as pd
# create first DataFrame
df1 = pd.DataFrame({
'A': ['A0', 'A1', 'A2', 'A3'],
'B': ['B0', 'B1', 'B2', 'B3']
}, index=['K0', 'K1', 'K2', 'K3'])
# create second DataFrame
df2 = pd.DataFrame({
'C': ['C0', 'C1', 'C2', 'C3'],
'D': ['D0', 'D1', 'D2', 'D3']
}, index=['K0', 'K1', 'K2', 'K3'])
# join two DataFrame
result = df1.join(df2)
print(result)Output result:
A B C D
K0 A0 B0 C0 D0
K1 A1 B1 C1 D1
K2 A2 B2 C2 D2
K3 A3 B3 C3 D3Using the indicator parameter of merge to track the source of the merge
pd.merge(df1, df2, on='key', how='outer', indicator=True)- Example code
import pandas as pd
# create first DataFrame
df1 = pd.DataFrame({
'key': ['K0', 'K1', 'K2', 'K3'],
'A': ['A0', 'A1', 'A2', 'A3'],
'B': ['B0', 'B1', 'B2', 'B3']
})
# create second DataFrame
df2 = pd.DataFrame({
'key': ['K0', 'K1', 'K2', 'K3'],
'C': ['C0', 'C1', 'C2', 'C3'],
'D': ['D0', 'D1', 'D2', 'D3']
})
# merge two DataFrame
result = pd.merge(df1, df2, on='key')
print(result)Output result:
key A B C D
0 K0 A0 B0 C0 D0
1 K1 A1 B1 C1 D1
2 K2 A2 B2 C2 D2
3 K3 A3 B3 C3 D3Using concat for Axial Merging
pd.concat([df1, df2], axis=1, keys=['2024', '2025'])- Vertical concatenation (default): Concatenate multiple DataFrames in the row direction. Columns with the same name will be merged.
- Horizontal concatenation: By setting
axis=1, you can concatenate multiple DataFrames in the column direction. Rows with the same index will be merged. - Keeping source identifiers: Using the
keysparameter, you can add identifiers to each original DataFrame to generate a multi-level index. - Handling non-overlapping columns:
By default,join='outer'is used to keep all columns, and missing values are filled with NaN.
Settingjoin='inner'will only keep the columns that are common to all DataFrames. - Index handling:
By default, the original index is retained, which may lead to duplicates.
Settingignore_index=Truecan generate a new continuous integer index.
import pandas as pd
# Create sample DataFrames
df1 = pd.DataFrame({
'A': ['A0', 'A1', 'A2', 'A3'],
'B': ['B0', 'B1', 'B2', 'B3']
}, index=[0, 1, 2, 3])
df2 = pd.DataFrame({
'A': ['A4', 'A5', 'A6', 'A7'],
'B': ['B4', 'B5', 'B6', 'B7']
}, index=[4, 5, 6, 7])
df3 = pd.DataFrame({
'C': ['C0', 'C1', 'C2', 'C3'],
'D': ['D0', 'D1', 'D2', 'D3']
}, index=[0, 1, 2, 3])
# Example 1: Vertical concatenation (default axis=0)
vertical_concat = pd.concat([df1, df2])
print("Example 1 - Vertical concatenation result:")
print(vertical_concat)
# result:
'''
A B
0 A0 B0
1 A1 B1
2 A2 B2
3 A3 B3
4 A4 B4
5 A5 B5
6 A6 B6
7 A7 B7
'''
# Example 2: Horizontal concatenation (axis=1)
horizontal_concat = pd.concat([df1, df3], axis=1)
print("\nExample 2 - Horizontal concatenation result:")
print(horizontal_concat)
# result
'''
A B C D
0 A0 B0 C0 D0
1 A1 B1 C1 D1
2 A2 B2 C2 D2
3 A3 B3 C3 D3
'''The raw data obtained by the crawler can be directly stored in databases such as MySQL and MongoDB after the above data cleaning.
df.to_sql('table_name',con)At this time, the data stored in the database no longer contains null values, duplicate values, or dirty data that does not meet the format.
